Project details
RosettaLigand
Company: Vanderbilt University
Period: 2008 - 2013
Duration: 5.8 years (full-time equivalent)
- C++
- Computational Biology
- Python
Rewrote RosettaLigand to allow simultaneous docking of multiple ligands, cofactors and ions. Added functionality to allow large flexible ligands to be broken into small fragments. Fragment rotamers are used to sample ligand flexibility, analogous to protein side-chain rotamers.
Developed RosettaLigandDesign, software for designing small molecules within a flexible protein receptor. The protocol can simulate combinatorial chemistry as well as fragment extension. Built a fragment library based on structures seen in the Cambridge Structural Database.
About me
Senior Software Developer at University of Utah
My skills
- JavaScript
- Java
- Python
- C++
- jQuery
- Machine Learning
- Bioinformatics
- Computational Biology
Endorse Gordon's project
See Gordon's profile
All Gordon's projects
-
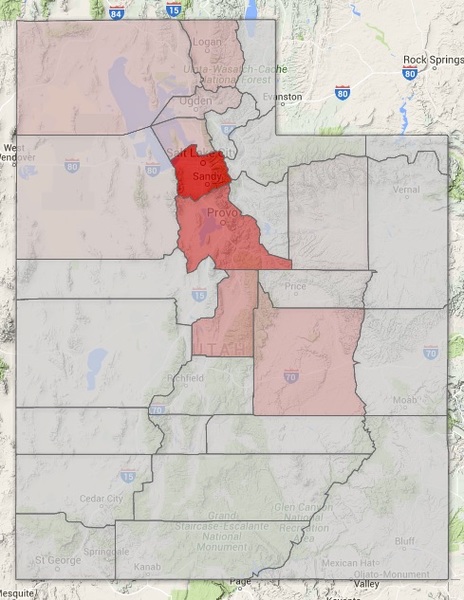
Learning from the Utah Population Database
(click to see)
2014 - Current (2.0 years FTE)
Client: University of Utah, Dept of Human Genetics
Industry: Science
- Bioinformatics
- Java
- JavaScript
- Python
- jQuery
Rewrote RosettaLigand to allow simultaneous docking of multiple ligands, cofactors and ions. Added functionality to allow large flexible ligands to be broken into small fragments. Fragment rotamers are used to sample ligand flexibility, analogous to protein side-chain rotamers.
Developed RosettaLigandDesign, software for designing small molecules within a flexible protein receptor. The protocol can simulate combinatorial chemistry as well as fragment extension. Built a fragment library based on structures seen in the Cambridge Structural Database.
-
Cancer genome analysis / variant calling
(click to see)
2013 - 2014 (1.2 years FTE)
Client: Saint Jude Children's Research Hospital
Industry: Science
- Bioinformatics
- Computational Biology
- Java
- Python
Rewrote RosettaLigand to allow simultaneous docking of multiple ligands, cofactors and ions. Added functionality to allow large flexible ligands to be broken into small fragments. Fragment rotamers are used to sample ligand flexibility, analogous to protein side-chain rotamers.
Developed RosettaLigandDesign, software for designing small molecules within a flexible protein receptor. The protocol can simulate combinatorial chemistry as well as fragment extension. Built a fragment library based on structures seen in the Cambridge Structural Database.
-
RosettaLigand
(click to see)
2008 - 2013 (5.8 years FTE)
Client: Vanderbilt University
Industry: Pharmaceutical
- C++
- Computational Biology
- Python
Rewrote RosettaLigand to allow simultaneous docking of multiple ligands, cofactors and ions. Added functionality to allow large flexible ligands to be broken into small fragments. Fragment rotamers are used to sample ligand flexibility, analogous to protein side-chain rotamers.
Developed RosettaLigandDesign, software for designing small molecules within a flexible protein receptor. The protocol can simulate combinatorial chemistry as well as fragment extension. Built a fragment library based on structures seen in the Cambridge Structural Database.
-
Real time PCR primer/probe sensitivity/specificity analysis
(click to see)
2007 - 2007 (16 weeks FTE)
Client: Lawrence Livermore National Laboratory / DHS
Industry: Science
Rewrote RosettaLigand to allow simultaneous docking of multiple ligands, cofactors and ions. Added functionality to allow large flexible ligands to be broken into small fragments. Fragment rotamers are used to sample ligand flexibility, analogous to protein side-chain rotamers.
Developed RosettaLigandDesign, software for designing small molecules within a flexible protein receptor. The protocol can simulate combinatorial chemistry as well as fragment extension. Built a fragment library based on structures seen in the Cambridge Structural Database.
-
Characterization of Xenopus tropicalis gene expression during development
(click to see)
2006 - 2006 (20 weeks FTE)
Client: Lawrence Livermore National Laboratory
Industry: Science
- Bioinformatics
Rewrote RosettaLigand to allow simultaneous docking of multiple ligands, cofactors and ions. Added functionality to allow large flexible ligands to be broken into small fragments. Fragment rotamers are used to sample ligand flexibility, analogous to protein side-chain rotamers.
Developed RosettaLigandDesign, software for designing small molecules within a flexible protein receptor. The protocol can simulate combinatorial chemistry as well as fragment extension. Built a fragment library based on structures seen in the Cambridge Structural Database.